Antibiotic resistance is proving to be one of the most serious threats to human health. In 2019 alone, it claimed around 1.27 million lives directly and played a role in nearly five million additional deaths. The situation worsened during the COVID-19 pandemic, and what compounds the challenge is the absence of new antibiotic classes developed for an extended period.
Researchers have now reported that they discovered a new class of antibiotic candidates using artificial intelligence (AI). A team from James Collins' group at the Broad Institute of the Massachusetts Institute of Technology and Harvard University employed deep learning AI to screen millions of chemicals for antibiotic activity.
Related: What NOT to Eat While Taking Antibiotics
Upon testing 283 potential compounds on mice, the researchers found that many proved effective against methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant enterococci, two of the most difficult bacteria to kill. Unlike a normal AI model, which operates as an incomprehensible "black box," this model's logic and physiology could be followed.

“Our [AI] models tell us not only which compounds have selective antibiotic activity, but also why, in terms of their chemical structure,” says Felix Wong at the Broad Institute of MIT and Harvard in Massachusetts.
Wong and his colleagues wanted to push the boundaries of AI-guided medication discovery with their research project. Their goal was to show that this approach could go beyond identifying specific drug targets and into predicting the broader biological effects associated with entire classes of drug-like substances.
First, more than 39,000 chemicals were tested to see how they affected Staphylococcus aureus and three different human cell types derived from the liver, skeletal muscle, and lungs. The accumulated data was then used as a training dataset for artificial intelligence models, enabling them to better comprehend the different structural patterns in each compound's chemical atoms and bonds. Because of this in-depth information, the AIs were able to predict the antibacterial activity of the chemicals and assess their potential harm to human cells.
Trained AI models analyzed 12 million chemicals using computer simulations, uncovering 3,646 molecules with optimum drug-like properties. Following that, calculations were used to find the specific chemical substructures that could reveal the features of each compound. The researchers discovered novel classes of possible antibiotics by carefully comparing these substructures between various substances. Two non-toxic chemicals with activity against both MRSA and vancomycin-resistant Enterococci were found in this groundbreaking finding.
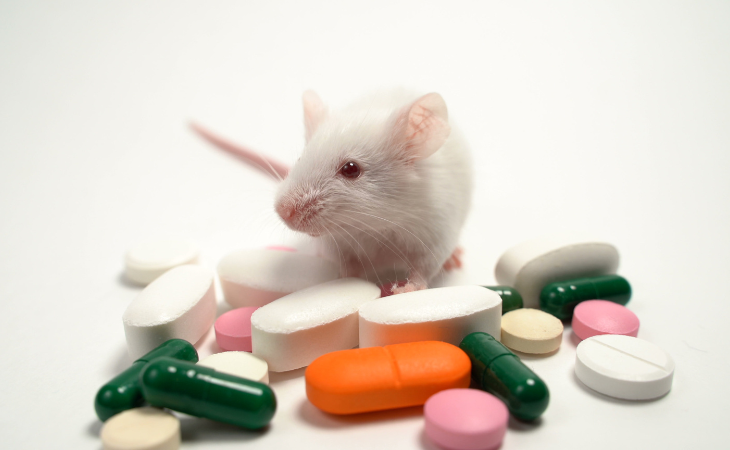
Finally, the researchers conducted mouse experiments to demonstrate the efficiency of these chemicals in treating MRSA-induced skin and thigh infections. According to James Collins, a co-author of the study from the Broad Institute, only a few recently found antibiotic families, such as oxazolidinones and lipopeptides, have shown effectiveness against both MRSA and vancomycin-resistant Enterococci. However, resistance to these chemicals is increasing.
“Our work identifies a new class of antibiotics, one of the few in 60 years, that complements these other antibiotics,” he says.
Related: It’s Essential that We Debunk These Antibiotic Myths!
Researchers are currently using this AI-guided technology to develop novel antibiotics and discover new drug categories. Compounds designed to specifically destroy aging and damaged cells implicated in illnesses such as osteoarthritis and cancer are among them.
The results of the study were published in the journal Nature.
 Go to BabaMail
Go to BabaMail